Psoriasis vulgaris and psoriatic arthritis are interrelated disorders with an important genetic component. While linkage studies have identified several candidate loci and genes, only recent technological advances and extensive genome-wide association studies have provided robust evidence of associations between psoriasis and several genes inside and outside the major histocompatibility complex. Most of these genes can be incorporated into an integrated pathogenic model of psoriatic disease comprising distinct signaling networks affecting skin barrier function (LCE3, DEFB4, GJB2), innate immune responses involving nuclear factor-κB signaling (TNFAIP3, TNIP1, NFKBIA, REL, FBXL19, TYK2, NOS2, CARD14), and adaptive immune responses involving CD8 T cells and interleukin 23 (IL-23)/IL-17-mediated lymphocyte signaling (HLA-C, IL12B, IL23R, IL23A, TRAF3IP2, ERAP1). A better understanding of the potential gene/gene and gene/environment interactions and of the functions of altered transcripts will undoubtedly have nosologic, therapeutic and prognostic implications.
La psoriasis vulgar y la artritis psoriásica son trastornos relacionados entre sí, con un importante componente genético. Aunque los estudios de ligamiento han llevado a la identificación de diversos loci y genes de susceptibilidad, ha sido el reciente progreso tecnológico y la realización de estudios de asociación genómica extensos lo que ha permitido demostrar asociaciones robustas de la psoriasis con diversos genes, asociados o no al complejo mayor de histocompatibilidad. La mayoría de estos genes se pueden incorporar en un modelo patogénico integrado que comprende distintas redes de señalización que afectan la función barrera de la piel (LCE3, DEFB4, GJB2), la respuesta inmune innata implicando al sistema de señales del factor nuclear-κB (TNFAIP3, TNIP1, NFKBIA, REL, FBXL19, TYK2, NOS2, CARD14), y la respuesta inmune adaptativa implicando a linfocitos T CD8 y las señales de la vía interleucina 23 (IL-23)/IL-17 (HLA-C, IL12B, IL23R, IL23A, TRAF3IP2, ERAP1). La mejor comprensión de las potenciales interacciones entre los genes implicados y de estos con factores ambientales, así como el conocimiento de las alteraciones en las funciones de las proteínas codificadas tendrán sin duda implicaciones nosológicas, terapéuticas y pronósticas.
Evidence suggests that psoriasis vulgaris is not a genetically homogenous disease but rather several different disease phenotypes associated with different genetic variants. For example, purely palmoplantar pustulosis can be considered a separate entity from psoriasis vulgaris, which in turn is genetically more closely associated with guttate psoriasis.1 Psoriasis vulgaris is a chronic inflammatory disease that shows a clear association with certain alleles of the HLA-C gene, and specifically with the HLA-Cw6 allele (known as HLA-Cw*0602 when identified through high-resolution genotyping), present in 30% of psoriasis patients (compared with between 10% and 15% in the general population). The relative risk of developing the disease is 2.5 greater in homozygous individuals than in heterozygous ones.2HLA-Cw6-positive patients have certain clinical characteristics defined by an early onset of the disease, presence of more extensive plaques, and a higher incidence of the Koebner phenomenon. In addition, more frequent streptococcal throat infection and high sensitivity to sunlight may be trigger factors and markers of more severe disease. HLA-Cw6-negative patients, in contrast, have a higher frequency of nail disorders and psoriatic arthritis.2,3
Before detailing the most important genetic aspects of the disease, we will review the underlying immunopathogenic mechanisms to enable a more integrated overview of the major therapeutic advances in recent years. These advances will then be described in detail.
Immunopathogenesis of PsoriasisPsoriasis is characterized by marked epidermal proliferation and abnormal differentiation with immune activation of keratinocytes, accompanied by numerous inflammatory and immune disorders, in which both innate and acquired immunity participate.4–6 The efficacy of cyclosporin, demonstrated more than 25 years ago, pointed to the fundamental role of T-lymphocytes.7 Subsequently, the efficacy of selective T-cell modulators further confirmed the importance of these types of cells.8–10 The marked efficacy of biologic agents that target tumor necrosis factor alfa (TNF-α) has also been demonstrated recently.11 This cytokine acts as a pleiotropic mediator of different types of inflammation. Likewise, anti-p40 antibodies, which block differentiation and expansion of Th1 and Th17 lymphocytes through interleukin (IL) 12 and IL-23, respectively, have also been found to be effective.12
The role of TNF-α and skin-resident T-lymphocytes has been confirmed in an experimental model with AGR129 mice, which lack the genes that encode interferon (IFN) and natural-killer (NK) cells, and so are unable to reject human skin.13 When apparently healthy skin from patients with psoriasis was grafted onto these mice, they spontaneously developed psoriasis plaques without the addition of CD4+ T lymphocytes. Serial biopsies showed that human T lymphocytes resident in the grafts proliferate and produce TNF-α, and treatment with human anti-CD3 antibodies, which impede T-lymphocyte proliferation, or TNF-α inhibitors (infliximab or etanercept), prevented conversion of the prepsoriatic skin into psoriasis lesions.13 Moreover, blockade of the exocytosis of T lymphocytes to the epidermis with an anti-integrin α1β1 antibody limits lesion development. When T lymphocytes are already present in the epidermis, inhibition is partial, and treatment is ineffective when fully developed psoriasis lesions are grafted, thereby confirming the role of resident T lymphocytes and their migration to the epidermis in the development of psoriasis lesions.14
The recent discovery of a subpopulation of T lymphocytes that express IL-17, and whose expression is determined by the action of IL-23 produced by antigen-presenting cells and dendritic cells on naïve T-cell precursors,15 has greatly changed our understanding of the pathogenesis of psoriasis. A marked expansion of cytotoxic T lymphocytes, which independently express IL-17 and IL-22 in the psoriatic epidermis, has been reported.16,17 The expansion of Th1 lymphocytes feeds back into this process by stimulating synthesis of IL-12 and IL-23 by antigen-presenting cells through production of IFN-γ.16
Genetic Linkage Studies of PsoriasisIn the 1990s, several groups started gene linkage studies which analyzed the cosegregation of microsatellite genetic markers in families with members with psoriasis. At least 6 loci of susceptibility to psoriasis, denoted PSORS1 through PSORS6, were identified.18–21 The main genetic determinant of psoriasis (PSORS1), located in the 6p21 chromosomal region, accounts for between 30% and 50% of genetic susceptibility to the disease and probably corresponds to the HLA-Cw*0602 allele, although the determinant is not associated with cases of late-onset psoriasis.20 This allele has been postulated to allow the presentation of a putative epitope present in type-I keratins, specifically, those whose expression is upregulated in psoriasis. This epitope may act as an autoantigen, present cross-reactivity with streptococcal protein M, and perpetuate autoimmune response (and perhaps CD4+ response with NK receptors) mediated by CD8+ cells, which are able to recognize the major histocompatibility complex (MHC) class I, leading to chronic lesions.19 The second locus of susceptibility to psoriasis, PSORS2, identified through linkage studies in different families, is located in the 17q24-q25 region, where a susceptibility locus has also been identified for atopic dermatitis, and the putative genes are implicated in regulating the immunological synapsis.3,21 Linkage studies in families from different geographic regions have identified other loci (Table 1), such as PSORS3 (4q34), PSORS4 (1q21), PSORS5 (3q21), PSORS6 (19p13), PSORS7 (1p32), and PSORS9 (4q31), while others are under investigation.21,22 Linkage with PSORS1 is clearly the most important, however. Recently, the HLA-Cw6 allele has been confirmed as the locus responsible for the association in PSORS1,23 and this finding has been confirmed in an extensive study of an ethnically diverse population.24
Loci Associated With Psoriasis (PSORS) and Psoriatic Arthritis (PSORSA).
| Locus | Region | OMIM | Candidate Genes/Function |
| PSORS1 | 6p21.3 | 612410 | HLA-Cw6 |
| PSORS2 | 17q25.5-qter | 607211 | CARD14 |
| PSORS3 | 4q34 | 601454 | IRF-2 |
| PSORS4 | 1q21 | 603935 | Loricrin, filaggrin, Pglyrp3,4; S100 and late cornified envelope genes (in the epidermal differentiation complex) |
| PSORS5 | 3q21 | 604316 | SLC12A8, cystatin A, zinc finger protein 148 |
| PSORS6 | 19p13 | 605364 | JunB |
| PSORS7 | 1p | 605606 | PTPN22 (1p13), IL23R (1p32.1-31.2) |
| PSORS8/PSORSA1 | 16q | 610707 | CX3CL1, CX3R1, NOD2/CARD15 |
| PSORS9 | 4q31 | 607857 | IL15 |
| PSORS10 | 18p11 | 612410 | |
| PSORS11 | 5q31-q33 | 612599 | IL12B |
| PSORS12 | 20q13 | 612950 | ZNF313/RNF114, ubiquitin ligase |
| PSORS13 | 6q21 | 614070 | TRAF3IP2 |
Source: Adapted from Duffin et al.48
The recent development of genetic studies based on analyzing millions of polymorphisms in a single nucleotide (single nucleotide polymorphisms [SNP]) used as genetic markers, systematic mapping of human haplotypes, and the development of high performance genotyping platforms has enabled genome-wide association studies (GWAS). In GWAS, thousands or even millions of SNP markers are analyzed in each individual, such that they account for>90% of the common variation present in the human genome. Given that a large number of markers are analyzed, and often the genetic effects are moderate (odds ratio [OR]<2), these types of genetic study require large cohorts of patients and controls. In the case of psoriasis, the main loci defined by a genetic effect with an OR>1.25 are HLA-C, IL12B, IL23R, IL23A, IL4/IL13, TNFAIP3, TNIP1, TRAF3IP2, TYK2, and IFIH, although other loci are in the process of being identified and validated (Table 2). Some of these loci associated with psoriasis have also been found to confer susceptibility to other inflammatory diseases of an immune nature, and are suggestive in other cases of ethnic variations in the disease.25,26
Genes Associated With Psoriasis Not Included in the MHC (Generally Identified Through GWAS Using SNPs or Copy Number Variants).
| Candidate Gene | Region/Superposition With PSORS | OMIM | Proposed Function | Pleiotropism (Different Diseases With Which They Have Been Associated) |
| IL23R | 1p31.3 (PSORS7) | 607562 | Encodes the IL-23 receptor | Psoriasis, Crohn disease, ulcerative colitis, ankylosing spondylitis |
| IL12B | 5q33.3 | 161561 | Encodes the p40 subunit of IL-12 and IL-23 | Psoriasis, psoriatic arthritis |
| IL13 | 5q31.1 | 147683 | Encodes IL-13; near IL-4, IL-5, and the RAD50 complex | Psoriasis, psoriatic arthritis, asthma, atopy |
| IL23A | 12q13.3 | 605580 | Encodes the p19 subunit of IL-23 | Psoriasis, psoriatic arthritis |
| TNFAIP3 | 6q23.3 | 191163 | Encodes the A20 protein, which acts through ubiquitin, inhibiting the proinflammatory activation of TNF-α induced NFκB | Psoriasis, psoriatic arthritis, Crohn disease, rheumatoid arthritis, systemic lupus erythematosus, type 1 diabetes, celiac disease |
| TNIP1 | 5q33.1 | 607714 | Encodes the ABIN-1 protein, which reduces the proinflammatory activation of TNF-α-induced NFκB | Psoriasis, psoriatic arthritis |
| TRAF3IP2 | 6q21 | 607043 | Encodes a protein that disrupts IL-17 signaling and interacts with different members of the family of Rel/NF-κB transcription factors. | Psoriasis, psoriatic arthritis |
| ZNF313/RNF114 | 20q13 (PSORS12) | 612451 | Encodes a ubiquitin ligase that is expressed in the skin | Psoriasis |
| ADAM33 | 20p13 | 607114 | Disintegrin and metalloprotease 33 | Psoriasis, asthma |
| PTPN22 | 1p13.2 (PSORS7) | 600716 | Tyrosine phosphatase that participates in the signaling of T lymphocyte receptors | Psoriasis, rheumatoid arthritis, systemic lupus erythematosus |
| CDKAL1 | 6p22 | 611259 | Encodes protein kinase homologous protein | Psoriasis, Crohn disease, type 2 diabetes |
| KIR2DS1, KIR2DL1 | 19q13.4 | 604952, 604936 | Encode receptors similar to immunoglobulin that bind to HLA-C and regulate the NK cell response. | Psoriasis, psoriatic arthritis |
| LCE3D/LCE3A LCE3C_LCE3B_del | 1q21 (PSORS4) | 612616, 612613 | Encode late cornified envelope (LCE) proteins, highly expressed in psoriasis | Psoriasis |
| DEFB4 | 8p23.1 | 602215 | Encodes human β-defensin | Psoriasis |
| IL15 | 4q31.2-q32.1 (PSORS9) | Encodes an interleukin that affects the activation and proliferation of T lymphocytes | Psoriasis | |
| IL2, IL21 | 4q27 | 147680, 605384 | Encode interleukins that participate in the proliferation of T lymphocytes, Th17 differentiation, and keratinocyte proliferation | Psoriatic arthritis, rheumatoid arthritis, type 1 diabetes, ulcerative colitis |
| IL28RA | 1p36.11 | 607404 | Encodes the alfa subunit of the IL-23 receptor | Psoriasis |
| REL | 2p16.1 | 164910 | Encodes an oncogene member of the Rel/NFκB transcription factor family | Psoriasis |
| IFIH1 | 2q24.2 | 606951 | Encodes an interferon-induced helicase | Psoriasis |
| ERAP1 | 5q15 | 606832 | Encodes an aminopeptidase of the endoplasmic reticulum, participates in the processing of peptides by MHC-I | Psoriasis in patients positive for HLA-Cw*0602; early onset in Chinese individuals of the Han ethnicity |
| NFKBIA | 14q13.2 | 604495 | Encodes a protein that deactivates NFκB by sequestering it in the cytoplasm | Psoriasis |
| TYK2 | 19p13.2 | 176941 | Encodes a protein that participates in the signaling of the type I interferon receptor | Psoriasis, systemic lupus erythematosus |
| PTTG1 | 5q33.3 | 604147 | Participates in cell proliferation and transformation | Psoriasis |
| CSMD1 | 8p23.2 | 608397 | Product participates in complement activation (?) | Psoriasis |
| GJB2 | 13q12.11 | 121011 | Connexin 26 (keratoderma) | Psoriasis |
| SERPINB8 | 18q22.1 | 601697 | Protease 8 inhibitor (regulates multiple functions); increased expression in psoriasis lesions | Psoriasis |
| ZNF816A | 19q13.41 | Encodes a zinc finger protein (participates in the recognition of RNA and other proteins) of the same class as the ZNF313 gene product. | Early onset psoriasis in Chinese individuals of the Han ethnicity | |
| NOS2 | 17q11.2 | 163730 | Nitric oxide synthetase | Psoriasis, psoriatic arthritis, hypertension, malaria |
| FBXL19 | 16p11.2 | 609085 | Ubiquitin ligase | Psoriasis, psoriatic arthritis |
| PSMA6 (?) | 14q13.2 | 602855 | Proteasome subunit (regulates inflammation through NFκB) | Psoriasis, psoriatic arthritis, susceptibility to myocardial infarction |
| CARD14 | 17q25.3-qter | 607211 | Activation of NFκB; participates in apoptosis |
The corresponding references are given in the text.
Abbreviations: GWAS, genome wide association studies; MHC, major histocompatibility complex; SNP, single nucleotide polymorphism.
GWAS have provided genetic evidence of the implication of the IL-23 pathway in psoriasis. The first large-scale study of genetic association in psoriasis (which was not a GWAS in the strict sense because it only analyzed SNPs within or close to genes, but not in other regions of the genome) enabled identification of an SNP located in the 3’ terminal region of the IL12B gene. This gene encodes the p40 subunit common to IL-12 and IL-23, and was the first locus clearly and reproducibly associated with psoriasis risk and that was independent of major histocompatibility complex (MHC).27 A subsequent study has managed to identify a second polymorphism independently associated with the disease. In turn, 2 other SNPs have been identified in the locus that encodes one of the subunits of the IL-23 receptor (IL23R), and that also shows an independent association with psoriasis.28 The association of these loci has been validated in different population groups,29–31 both in psoriasis and in psoriatic arthritis.32,33 Likewise, the IL23A gene, which encodes the p19 subunit of IL-23, has been shown to be associated with psoriasis and psoriatic arthritis28 as well as with ankylosing spondylitis and Crohn disease, but not with rheumatoid arthritis and celiac disease.33
The second GWAS published confirmed the association of genes IL12B and IL23R with psoriasis and also with psoriatic arthritis.34 The study identified a new signal close to the IL23RB2 gene, as well as new candidate loci in the 13q13 region, which contains the COG6 and LHFP genes; the 15q21 region, which contains the TNFAIP8L3 gene; the 4q27 region, which contains the IL2 and IL21 genes; and the 1q21 region, which contains the LCE1C gene. However, the small sample size used in this study did not permit unequivocal association of these genes with psoriasis.
A GWAS, with analysis of both SNP and copy number variants (CNV), conducted in a large Chinese population of Han and Uygur ancestry,35 identified a new association with the cluster of genes of the late cornified envelope (LCE) on chromosome 1q21, previously identified as PSORS4 through gene linkage studies. The products encoded by the genes in this region participate in the terminal differentiation of the epidermis, making these genes excellent candidates to explain the different phenotypes of psoriasis. A study published at the same time identified a CNV also associated with psoriasis in this same chromosomal region. This CNV is a deletion (that is, a DNA segment is absent) and it correlates strongly (that is, there is a linkage imbalance) with SNP rs4112788 but not with SNP rs6701216, as published by Liu et al.34 The association of SNP correlated with LCE3C_LCE3B-del was confirmed in a population of British patients with psoriatic arthritis,36 but not in a population of German patients.37 In this case, the phenotypic heterogeneity of each cohort and the small sample size might explain the differences observed.
Genes do not act in isolation but operate through complex molecular networks and participate in different cellular pathways. Likewise, the association of certain genetic variants with the risk of developing the disease may be conditioned by the presence of other variants within the genome. Gene interaction, or epistasis, is a complex genetic mechanism, and until recently there was little evidence that such processes were operating in humans. It is relevant to note that the main evidence for the existence of epistasis in human diseases comes from psoriasis. In a Chinese population, the presence of epistatic interactions between the MHC and other risk genes, such as LCE and IL12B, was identified.38 The risk of psoriasis increases 26-fold in individuals with the risk alleles in MHC and LCE and 36-fold in individuals with risk alleles in MHC and IL12B, compared to individuals who are not carriers. However, in a study conducted in a population from the north of China, the investigators observed that association with LCE3C_LCE3B-del depends on the age of onset and the family history of the patient, and epistasis (modification of susceptibility) with the HLA-Cw6 allele was not detected.39
In 2006, investigators at the University of Michigan, the University of Washington in Saint Louis, and the University of Utah initiated a collaboration to carry out GWAS in psoriasis. In 2009, the consortium published the findings of the first GWAS, known as the Collaborative Association Study of Psoriasis (CASP). In the study, 438 670 SNPs from 1409 cases and 1436 controls were genotyped in a first phase, which was followed by genotyping of the 21 SNPs with strongest statistical evidence of an association, corresponding to 18 loci, in an independent cohort of 5048 cases and 5041 controls.28 For 10 loci, the study found evidence of association (P<.05 in the follow-up cohort) which was especially convincing (P<.0005 in the follow-up cohort) for 7 of these, confirming the association with HLA-Cw6, IL13, IL12B, and IL23R, and identifying the aforementioned association with IL23A. A novel finding of this study was the association of 2 genes with the signaling factor pathway for the transcription factor NF-κB (implicated in the pathogenesis of autoimmune diseases such as systemic lupus erythematosus and rheumatoid arthritis): TNF-α-induced protein 3 (TNFAIP3) and TNFAIP3-interacting protein 1 (TNIP1). The study did not find differences in terms of associations with psoriatic arthritis, unlike other studies in which IL13 seems to be specifically associated with psoriatic arthritis.35,40 Recently, investigators have published several GWAS in independent populations that confirm the association of the TRAF3IP2 gene with both psoriasis and psoriatic arthritis.41,42 This gene encodes a protein that disrupts IL-17-induced signal and interacts with different members of the family of Rel/NF-κB transcription factors.
Other signals have been repeatedly detected in different populations through GWAS or SNP.43–45 Examples include a locus close to the ZNF313/RNF114 gene, on 20q13, which, like TNFAIP3 and TNIP1, encodes a ubiquitin ligase,46 or the regions of the CDKAL1, PTPN22, and ADAM33 genes,47 which have been confirmed in the case of the former.26 This locus is also implicated in susceptibility to type 2 diabetes mellitus and Crohn disease.48,49
One of the most recent studies to be published reinforces the hypothesis that the pathogenesis of psoriasis combines genetic determinants of epidermal barrier dysfunction with disrupted regulation of innate and adaptive immunity. The Genetic Analysis of Psoriasis Consortium and Wellcome Trust Case Control Consortium 2 conducted a GWAS with 594 224 SNPs in 2622 patients with psoriasis and 5667 controls. The association with TRAF3IP2 was confirmed, and 7 new loci that contained genes with immune function were identified50: IL28RA, REL, IFIH1, ERAP1, NFKBIA, and TYK2. These associations were validated in a replication cohort of 9079 samples from European Caucasian individuals.51 The study identified the epistatic association of the HLAC and ERAP1 genes with the risk of developing psoriasis. The ERAP1 gene product participates in the processing of peptides by class i MHC and the risk allele for this gene only increases the risk of psoriasis in those individuals who are positive for the HLA-Cw*0602 allele.51 This is one of the first clear and reproducible examples of epistasis in humans.
A recent GWAS replication study,52 performed in China,37 and including 8312 patients with psoriasis and 12 919 Chinese controls, 3293 cases and 4188 controls in German and the United States, and 254 nuclear families in the United States, identified 6 new susceptibility loci that contained gene candidates ERAP1, PTTG1, CSMD1, GJB2, SERPINB8, and ZNF816A, which replicated a locus on 5q33.1 (TNIP1-ANXA6), previously reported in European studies. Two of the loci identified (ZNF816A and GJB2) also show evidence of an association in a study of the German population. Moreover, ERAP1 and ZNF816A are associated with type 1 psoriasis (that is, early-onset psoriasis) in Chinese individuals of Han ancestry. Apart from identifying new factors of genetic susceptibility, this study clearly illustrates that part of the genetic heterogenicity present in the disease can be attributed to genetic differences between ethnically different populations.
The results of different GWAS can be pooled to increase statistical power and thus identify new risk loci. Recently, a metaanalysis of 2 of the aforementioned GWAS and a more recent third GWAS conducted in a new cohort of 1831 cases and 2546 controls and replicated in 4064 cases and 4685 controls in Michigan, Toronto, Terranova, and Germany,53 identified 3 new susceptibility loci, NOS2, FBXL19, and one near PSMA6-NFKBIA. All these were associated with both cutaneous psoriasis and psoriatic arthritis. Likewise, in this study, the association of a signal near RNF114, described recently, was replicated.49,51
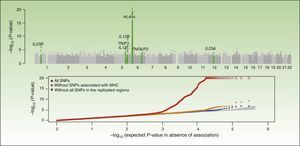
Integrative Concept of the Genetic Component and Immunopathogenesis of PsoriasisAs illustrated by the results of the CASP study (Figure 1), the MHC is the main determinant of genetic susceptibility in psoriasis. Several SNPs in the region of the MHC have been associated with psoriasis in different GWAS. The SNP with the strongest association, rs1219187, shows a marked linkage imbalance with the HLA-Cw*0602 allele,28 the main risk allele,54 but other independent signals are present such as rs2022544,26 rs2073048, located on c6orf10, a potential mediator in the TNF-α pathway, and rs13437088, at 30kb55 from HLA-B in the centromere direction and at 16kb from MICA (MHC class I polypeptide-related sequence A precursor) in the telomere direction.54 A detailed analysis in 2 Chinese populations of Han ancestry has identified an association of HLA-B*57 with an increased risk of psoriasis, and of HLA-B*40 with a decreased risk of disease, independently of HLA-Cw*0602 and the C6orf10 locus.54 The second association was validated recently in a study that showed that MICA*016 increased the risk of developing psoriasis without arthritis, and homozygosity for MICA*00801 increased the risk of developing the arthritic form of the disease in patients with psoriasis.56
Summary of the results of the Genome Wide Association Study known as the Collaborative Association Study of Psoriasis. The top plot shows the statistically significant values in relation to the chromosomal position. This type of plot is known as a Manhattan plot, as the highly significant regions resemble the skyline of a city with skyscrapers. In this case, the replication studies confirmed the association of 7 regions marked in the green plot. The lower plot, known as a QQplot, orders the values by significance (that is, observed P value) and compares them with the theoretical distribution in absence of an association (that is, expected P value). Such a plot readily reveals the existence of single nucleotide polymorphism (SNPs) associated with the disease as, in absence of any association, the values appear on the diagonal line. In this case, we see how the QQplot of the SNP of the human leukocyte antigen (HLA) region (in red) deviates clearly. When this region (orange) and the other associated regions (blue) are excluded, we see how the plot approaches the expected value (shaded zone). In both plots, the significance of the HLA-C region is truncated to facilitate interpretation of the results.
Source: Elder et al.26; Nair et al.28 Abbreviation: MHC, major histocompatibility complex.
Guttate psoriasis, which is associated with HLACw6 in up to 100% of cases,57 is often preceded by streptococcal throat infection (rarely by perianal dermatitis, balanoposthitis, or vulvovaginitis), and infrequently by other streptococcal infections of the skin such as impetigo and erysipelas. During the course of the throat infection, HLA-Cw6-mediated streptococcal antigens or superantigens are presented to the naïve tonsillar T lymphocytes that will proliferate and differentiate into an effector phenotype and a memory phenotype, and acquire a skin homing capacity (CLA+),26,54 whereas the peptidoglycan of the streptococcal wall could alternatively activate lymphocytes by activating cytokine-mediated Toll-like receptors.58 With time, oligoclonal expansion of the T lymphocytes directed against streptococcal antigens with skin homing will occur. These cells will begin to recognize epidermal autoantigens, giving rise to the plaque psoriasis lesions.59 The genetic predisposition determined by HLA-Cw6 and other components of the MHC may arise through loss of tolerance to epidermal autoantigens, especially if their expression is upregulated or they appear (neoantigens) in psoriasis. This is the case for a range of proteins, such as K16 and K17 keratins, β-defensin-4 (encoded by DEFB4), psoriasin (S100A7), calgranulin (S100A8 and S100A9), small proline-rich region proteins (SPRR), and CLE proteins, many of which are encoded in the epidermal differentiation complex located on 1q21.3 (PSORS4). Genes that encode human β-defensins (antimicrobial peptides with similar activity to cytokines) are located in several clusters. One of them located on the 8p23-1 region has recently been associated with an amplification-type CNV of the DEFB4, DEFB103, and DEFB104 genes, which encode the β-defensins 4, 3, and 2, respectively.60
Given that the vast majority of T lymphocytes in the psoriatic infiltrate do not show clonal expansion, other antigen-independent mechanisms must be in operation. In these mechanisms, HLA-C may also participate, acting as a ligand for the killer immunoglobulin-like receptors (KIR). These receptors regulate the activity of the NK-T cells and are encoded by the KIR gene, whose locus is associated with psoriasis and psoriatic arthritis.61–65
Another important genetic determinant of psoriasis presumably relates to regulation of the NF-kB signaling pathway (Figure 2). A20 and ABIN1, which are TNFAIP3 and TNIP1 gene products, respectively, participate in ubiquitin-mediated destruction of the IKKγ/NEMO complex and other components of the TNF-α signal transduction pathways. Recently, murine models have been developed with specific suppression of A20 expression in the skin and macrophages that develop psoriasiform dermatitis66 and arthritis.67 These experimental models confirm the importance of this pathogenic pathway in psoriasis and the role of A20 as an inhibitor of the activation of dendritic cells and autoimmune response.68,69
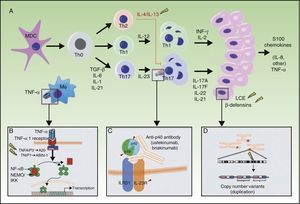
A, Main pathogenic pathways in psoriasis with genetic implications: the myeloid dendritic cells produce interleukin (IL)-12 and IL-23 after activation by different cytokines such as tumor necrosis factor (TNF)-α, interferon (IFN)-α, IFN-γ, and IL-6. Naïve T lymphocytes, in presence of tumor growth factor (TGF) β, IL-6, IL-1, and IL-21, differentiate into Th17 lymphocytes, which express the IL-23 receptor and proliferate in the presence of this cytokine. Th17 lymphocytes produce IL-17A, IL-17F, IL-22, and IL-21, which activate the keratinocytes in immunologic and proliferative terms, giving rise to the production of TNF-α, IL-1, IL-6, IL-8, S100A7, and other S100 proteins and antimicrobial peptides (β-defensins). A. The binding of TNF-α to its receptor activates a cascade of signals that give rise to the release of nuclear factor (NF)-κB from its inhibitory complex NFκB essential modulator/inhibitor of κB kinase (NEMO/IKK), leading to transcription of A20, a negative regulator of NFκB enhanced by the ABIN-1 protein. Psoriasis is associated with certain polymorphisms in the genes that encode these 2 inhibitory proteins. B. Inhibition of IL-23-mediated signaling is the mechanism of action of the p40 inhibiting monoclonal antibodies, such as ustekinumab. Psoriasis has also been associated with polymorphisms in the genes that encode the P19 and p40 subunits of IL-23 and IL-12/IL-23, respectively, as well as a subunit of the IL-23 receptor. C. An association has been reported between the CNVs of the LCE proteins and human β-defensins and psoriasis. Abbreviations: CNV, copy number variants; LCE, late cornified envelope; MDC, myeloid dendritic cells.
Source: Adapted from Duffin et al.45
Abnormal transcription of cytokines in the Th2 family (IL-13, IL-4, IL-5) may interfere with the negative regulation of differentiation of naïve T lymphocytes to Th17 lymphocytes70 and with IL-17 synthesis by these cells.71 Interestingly, although signals related to the IL-13 gene have been identified along with the cluster that regulates the transcription of different Th2 cytokines in different studies,48 this association has recently been found to be limited to patients with psoriatic arthritis.43
Recent AdvancesAs proof of the dynamic nature of genetic study in psoriasis, and by way of conclusion, we only need cite the 2 most recent advances in this field, which are related to monogenic forms of pustular psoriasis.
Generalized pustular psoriasis is characterized by repetitive episodes of generalized pustular rash accompanied by high fever, leukocytosis, and elevated levels of C reactive protein. Plaque psoriasis may also be present. A hereditary form has been reported with autosomal recessive transmission. A study of 9 Tunisian families with this disease identified linkage with a 1.2 megabase region on 2q13-q14.1 and a homozygous mutation in the IL36RH gene, which encodes the receptor antagonist for IL-36 (a cytokine with anti-inflammatory properties) and which is responsible for defective synthesis of a less stable and less potent variant in terms of interaction with IL1 receptor-like 2. This mutation leads to an increase in the production of IL-8 and other pro-inflammatory cytokines by keratinocytes, in turn leading to intraepidermal accumulation of polymorphonuclear molecules.72 Other mutations of the same gene have been identified in 3 sporadic cases of the same disease,73 which has become known as deficiency of the IL-36R antagonist (DITRA) and which bears strong resemblance to deficiency of the IL-1R antagonist (DIRA), an autoinflammatory disease reported in 2009 with strong response to treatment with IL-1 antagonists such as anakinra.74
Another significant advance was the identification of the CARD14 gene as responsible for the association with PSORS2 when mutations leading to increased function of the transcribed protein (caspase recruitment domain-containing protein 14) were found in 2 extended families, one in Europe with multiple cases of psoriasis and psoriatic arthritis in 30% of them and another in Taiwan, as well as a de novo mutation in a girl with early-onset sporadic pustular psoriasis.75 In another article, published by the same group, 15 additional variants of CARD14 are described along with their distribution in 7 cohorts of patients with psoriasis (more than 6000 cases and controls). These variants are suggestive of epistasis with HLA-Cw6 and their effect on activation of NF-κB and the transcription of different genes in keratinocytes transfected with different mutants.76 Caspase recruitment domain-containing protein 14 is usually localized to the basal and suprabasal layers of the epidermis, whereas expression in psoriatic lesions is upregulated in diffuse and localized fashion in the suprabasal layers. CARD14 mutations associated with the development of psoriasis lead to increased activation of NF-κB and the expression of different genes associated with psoriasis in keratinocytes.
In both cases, these findings reinforce the current hypothesis for pathogenesis linked to the role of keratinocytes in psoriasis, and extend our knowledge of the mechanisms of production of pustular lesions, as well as the exceptional monogenic forms of the disease.
Ethical ResponsibilitiesProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this investigation.
Confidentiality of dataThe authors declare that patient data do not appear in this article.
Right to privacy and informed consentThe authors declare that patient data do not appear in this article.
FundingThis manuscript was drafted as part of the Strategic Project PSE-010000-2006-6, funded by the Ministry of Science and Innovation.
Conflicts of InterestThe authors declare that they have no conflicts of interest.
Please cite this article as: Puig L, Julià A, Marsal S. Psoriasis: bases genéticas y patogenéticas. Actas Dermosifiliogr. 2014;105:535–545.