Numerous studies have investigated the association that exists between genetic variants and the efficacy profile of biologic therapies for the management of psoriasis. However, as far as we know, data on this association for brodalumab are lacking in the currently available scientific literature.
ObjectivesTo analyze the association of 180 polymorphisms with an optimal response to brodalumab in real-world clinical practice.
MethodsA total of 119 patients with plaque psoriasis on a 24-regimen of brodalumab recruited from 11 Spanish hospitals were genotyped for 180 polymorphisms. Optimal response was evaluated as absolute (PASI)≤1 at 6 and 12 months. Polymorphisms with false discovery rates<0.25 were included in a multiple regression model.
ResultsA total of 68% and 62% of patients achieved PASI≤1 at 6 and 12 months, respectively. Patient weight, history of biological therapy, disease-modifying anti-rheumatic drugs, and psoriatic arthritis were identified as risk factors for failing to achieve PASI≤1. At 12 months, polymorphisms rs495337 (SPATA2), rs6311 (HTR2A), and rs4085613 (LCE3D) were associated with achieving a PASI≤1 regardless of previous use of biologics and DMARDs, psoriatic arthritis, or weight. The genotypes CT-TT for rs6311 (HTR2A) and GT for rs4085613 (LCE3D) were identified as risk factors for lack of optimal response at 12 months, while genotypes AG-AA for rs495337 (SPATA2) increase the probability of response. No polymorphism was associated to brodalumab response at 6 months.
ConclusionsThis study identified genetic variations associated with the ability to achieve an optimal response to brodalumab, providing potential insights into its efficacy profile for treating plaque psoriasis.
No existen estudios sobre el uso de polimorfismos en psoriasis para predecir la respuesta a brodalumab.
ObjetivosAnalizar la relación de 180 polimorfismos con una respuesta óptima a brodalumab en práctica clínica real.
MétodosSe incluyeron pacientes con psoriasis en placas tratados con brodalumab durante al menos 24semanas. Se genotiparon 180 polimorfismos de genes relacionados con la inmunopatogenia de la psoriasis. La respuesta óptima (PASI absoluto ≤1) al tratamiento se evaluó a los 6 y 12meses. En el análisis multivariante se incluyeron los polimorfismos con False Discovery Rate ≤0,25.
ResultadosEl estudio incluyó 119 pacientes; el 68% y el 62% alcanzaron un PASI ≤1 a los 6 y a los 12meses, respectivamente. Se identificaron como factores de riesgo para no lograr un PASI ≤1: el uso previo de biológicos y fármacos antirreumáticos modificadores de la enfermedad, diagnóstico de artritis psoriásica y el peso. El análisis de asociación a los 12meses reveló que los polimorfismos rs495337 (SPATA), rs6311 (HTR2A) y rs4085613 (LCE3D) se asocian con alcanzar un PASI ≤1 independientemente del uso previo de biológicos, artritis psoriásica o el peso. Los genotipos CT-TT para rs6311 y GT para rs4085613 fueron identificados como factores de riesgo para alcanzar PASI ≤1, mientras que AG-AA para rs495337 aumentaron la probabilidad de lograr respuesta óptima. Ningún polimorfismo se asoció con la respuesta a los 6meses.
ConclusionesEste estudio identificó variaciones genéticas que podrían predecir la respuesta a brodalumab, permitiendo optimizar su uso en el tratamiento de la psoriasis.
Psoriasis is a multifactorial disease influenced by genetic predisposition, epigenetic changes, and environmental triggers. The development of psoriatic lesions results from the intricate interplay of these factors.1 Interleukin (IL)-17 is a key mediator in this process promoting inflammation and tissue damage. While IL-17 production was initially attributed to IL-23-dependent T helper(Th)17 cells, emerging evidence suggests that innate immune cells are a source of IL-23-independent IL-17.2
The approval of biologic therapies targeting the IL-23/IL-17 axis has provided treatments with an impressive efficacy profile in a substantial number of psoriasis patients.3 Brodalumab—a fully human monoclonal antibody—stands out among these novel therapies. By binding to IL-17 receptor subunit A (IL-17RA), brodalumab inhibits the activity of multiple pro-inflammatory cytokines of the IL-17 family, beyond IL-17A. Brodalumab is indicated for moderate-to-severe psoriasis in adults eligible for systemic therapy. Clinical trials with this drug have demonstrated highly favorable safety and efficacy outcomes leading to rapid control of the disease.4 However, real-world efficacy rates may vary,5 emphasizing the importance of identifying predictive markers for treatment response. Genetic markers, particularly single nucleotide polymorphisms (SNPs), are promising candidates.6
Numerous studies, mainly focues on anti-tumor necrosis factor (TNF) blockers and the anti-IL-17 monoclonal antibody ustekinumab, have looked into the association of genetic variations and the efficacy of biologic therapies in the management of psoriasis.7,8 In addition, we recently identified the association of certain variants with the efficacy profile of secukinumab.9 However, this association remains to be explored for brodalumab.
We studied the association of 180 polymorphisms in relevant genes for psoriasis with the response of psoriatic patients to brodalumab at 12 months in real-world clinical settings. The study focused on the association with patients with optimal response (super-responders) defined in this manuscript as those with absolute psoriasis area and severity index (PASI) scores≤1.
MethodsSubjectsWe conducted this retrospective-prospective study across 11 dermatology centers in Spain. The study was approved by Hospital Universitario de La Princesa Clinical Research Ethics Committee (Madrid, Spain). A total of 119 adult patients with moderate-to-severe chronic plaque psoriasis who had received prior treatment with brodalumab in real-world settings for, at least, 24 weeks were included. Patients were recruited between September 2020 and September 2022 and adhered to the dosage and therapeutic regimen recommended on the summary of product characteristics (210mg subcutaneously on weeks 0, 1, and 2, followed by doses every 2 weeks). Clinical data, age, sex, weight, disease duration, past medical history of systemic conventional and biological therapies, and presence of psoriatic arthritis (PsA) were collected. The efficacy profile of brodalumab was evaluated at 6 and 12 months and, then, categorized using absolute PASI scores ≤3, ≤2, and ≤ 1.
Sample processing and genotypingDNA extraction was performed using 1mL of peripheral blood with the MagNA Pure LC 2.0 system (Roche, Switzerland) and quantified using a NanoDrop® ND-1000 Spectrophotometer (Wilmington, United States). A customized microarray containing 180 polymorphisms in relevant genes for psoriasis was designed based on an extensive literature review (Supplementary Table 1).9 Genotyping was performed using a QuantStudio 12K Flex qPCR instrument with an OpenArray thermal block (Applied Biosystems, Thermofisher, United States).
StatisticsPolymorphisms with >5% missing genotypes and individuals with >5% missing data were excluded from the association analysis. Univariable logistic regression assessed the association of sociodemographic and clinical variables (age, sex, PsA, disease duration, and baseline PASI score) with treatment efficacy. The association between each polymorphism and clinical response was adjusted for covariates identified as significant in the univariate analysis using the Akaike Information Criterion (AIC) for model selection and the SNPassoc R package for analytical purposes. The Last Observation Carried Forward method was employed in the statistical analysis to impute missing data for patients who dropped out, primarily due to insufficient efficacy.
For the multivariable logistic regression model at 12 months, variants with a false discovery rate (FDR)<0.25 were selected. Model construction used the stepwise backward selection method. Receiver Operating Characteristic (ROC) curve analysis was used to assess the models’ ability to distinguish between responders and non-responders based on absolute PASI. Statistical analysis was performed using RStudio version 2023.09.0+463.
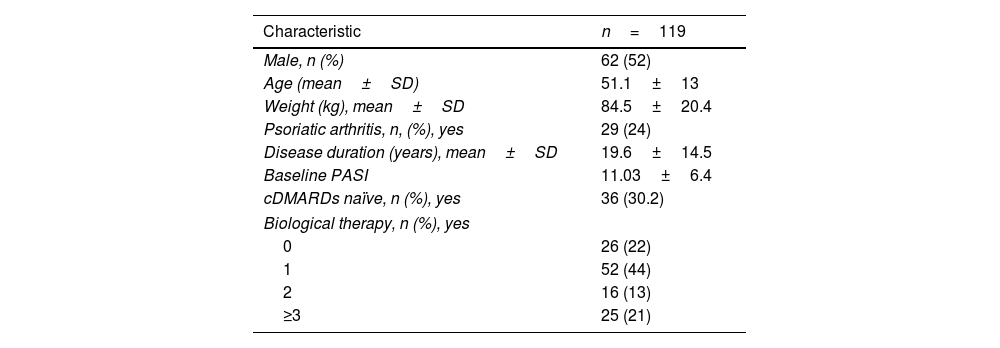
ResultsBaseline sociodemographic data and efficacy profileA cohort of 119 patients (52% men with a mean age of 51.1±13 years and a mean weight of 84.5±20.4kg) was analyzed. Among them, 22% were treatment-naïve to biological agents, and 24% had a confirmed diagnosis of PsA. The clinical and sociodemographic characteristics of the patients are shown in Table 1. No serious adverse events were reported.
Phenotypic characteristics of patients (baseline data).
| Characteristic | n=119 |
|---|---|
| Male, n (%) | 62 (52) |
| Age (mean±SD) | 51.1±13 |
| Weight (kg), mean±SD | 84.5±20.4 |
| Psoriatic arthritis, n, (%), yes | 29 (24) |
| Disease duration (years), mean±SD | 19.6±14.5 |
| Baseline PASI | 11.03±6.4 |
| cDMARDs naïve, n (%), yes | 36 (30.2) |
| Biological therapy, n (%), yes | |
| 0 | 26 (22) |
| 1 | 52 (44) |
| 2 | 16 (13) |
| ≥3 | 25 (21) |
Abbreviations: Data are shown as mean and standard deviation or number; PsA: psoriatic arthritis; PASI: psoriasis area and severity index; cDMARDs: conventional disease-modifying antirheumatic drugs.
Six months into treatment, 87%, 76%, and 68% of the patients achieved PASI≤3, ≤2, and ≤1, respectively. These favorable responses were maintained at 12 months; 87% of the patients maintained PASI≤3; 77%, PASI≤2; and 62%, PASI≤1.
Association studyFor the association analysis, a total of 5 patients were excluded due to genotyping failure. In the univariate analysis, prior use of disease-modifying anti-rheumatic drugs (DMARDs), prior use of biologics, PsA, and patient weight showed a significant association with optimal brodalumab response (PASI≤1,super-responders) (p-value<0.5). No association was ever found between sex, age, disease duration or baseline PASI, and treatment response.
At 6 months, several polymorphisms showed significant association (p<0.05) after adjusting for weight, DMARD use, prior biological treatment, and PsA. However, none of them met the cutoff for multiple corrections (FDR<0.25).
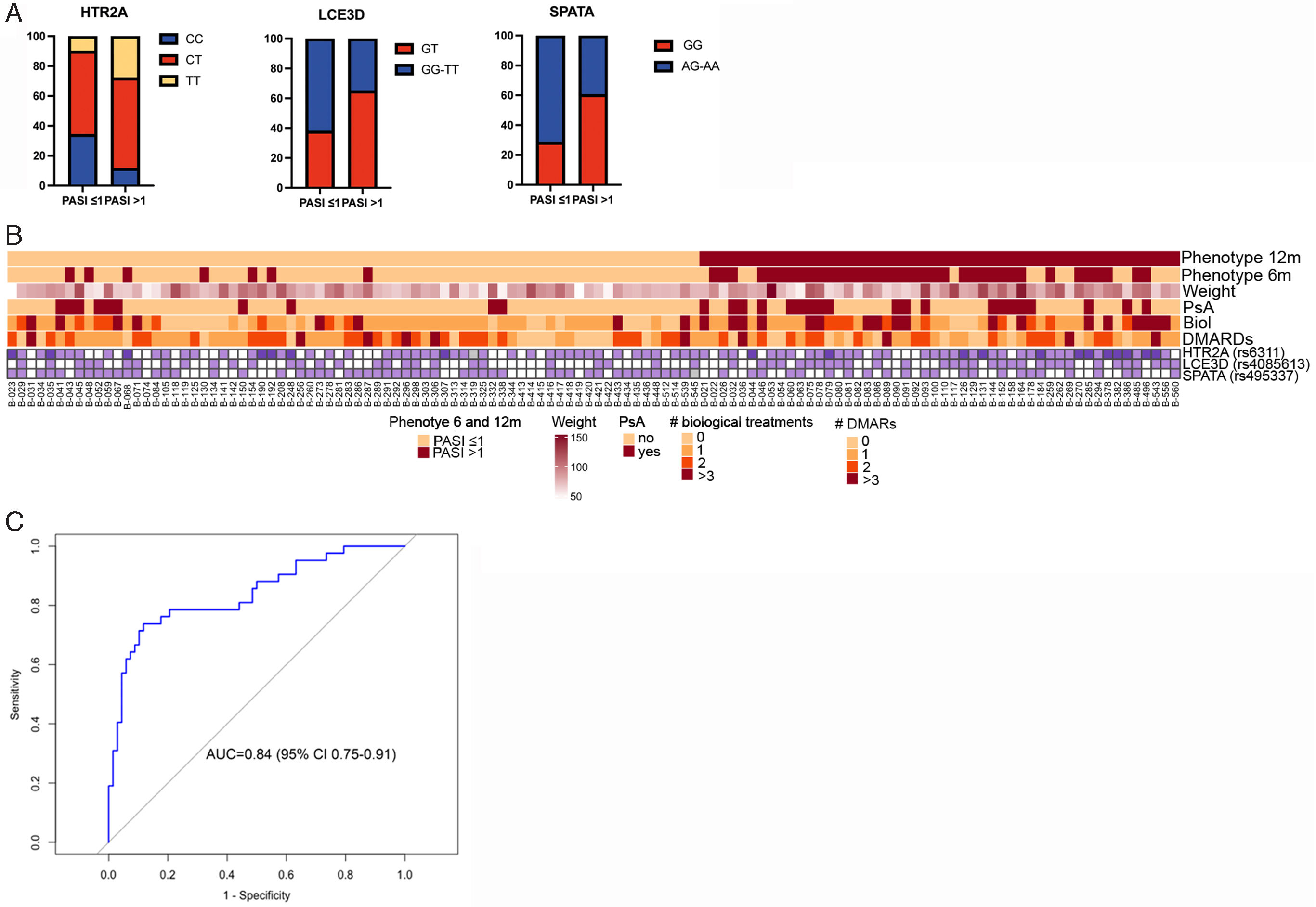
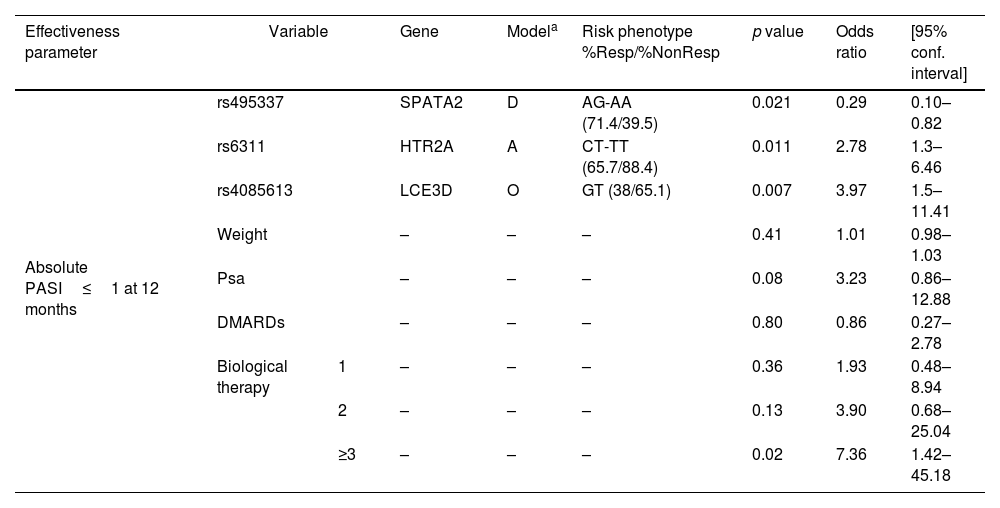
The 12-month analysis of treatment revealed that polymorphisms rs495337 (SPATA2), rs6311 (HTR2A), and rs4085613 (LCE3D) were associated with achieving absolute PASI≤1 regardless of the past medical history of use of biologic agents and DMARDs, PsA, or weight. The genotypes CT-TT for rs6311 (HTR2A) and GT for rs4085613 (LCE3D) were identified as risk factors for failure to achieve an absolute PASI≤1 at 12 months, while genotypes AG-AA for rs495337 (SPATA2) were protective (Table 2 and Fig. 1A, B). The ROC curve analysis revealed that the model including the combination of polymorphisms for SPATA2, HTR2A, and LCE3D, along with weight, previous biological and DMARD treatment, and presence of PsA, an area under the curve of 0.83 (95% confidence interval [CI], 0.75–0.91), with sensitivity and specificity rates of 0.69 and 0.91, respectively, for discrimination between super-responders and non- responders (Fig. 1C).
Multivariable logistic regression model for PASI≤1 at 12 months.
| Effectiveness parameter | Variable | Gene | Modela | Risk phenotype %Resp/%NonResp | p value | Odds ratio | [95% conf. interval] | |
|---|---|---|---|---|---|---|---|---|
| Absolute PASI≤1 at 12 months | rs495337 | SPATA2 | D | AG-AA (71.4/39.5) | 0.021 | 0.29 | 0.10–0.82 | |
| rs6311 | HTR2A | A | CT-TT (65.7/88.4) | 0.011 | 2.78 | 1.3–6.46 | ||
| rs4085613 | LCE3D | O | GT (38/65.1) | 0.007 | 3.97 | 1.5–11.41 | ||
| Weight | – | – | – | 0.41 | 1.01 | 0.98–1.03 | ||
| Psa | – | – | – | 0.08 | 3.23 | 0.86–12.88 | ||
| DMARDs | – | – | – | 0.80 | 0.86 | 0.27–2.78 | ||
| Biological therapy | 1 | – | – | – | 0.36 | 1.93 | 0.48–8.94 | |
| 2 | – | – | – | 0.13 | 3.90 | 0.68–25.04 | ||
| ≥3 | – | – | – | 0.02 | 7.36 | 1.42–45.18 | ||
Abbreviations: CI: confidence interval; OR: odds ratio; PASI: psoriasis area and severity index; Psa: psoriatic arthritis; DMARDs: drug modifying anti-rheumatic drugs; Resp: responders; Nonresp: non-responders.
Polymorphisms in HTR2A, LCE3D and SPATA associate with an optimal response (PASI≤1) to brodalumab at 12 months. (A) Genotypes of HTR2A, LCE3D and SPATAATG5 in patients achieving or not an absolute PASI≤1 at 12 months (percentages of patients with indicated genotype is shown). (B) Individual data from patients corresponding to polymorphisms in HTR2A, LCE3D and SPATA genes, weight, PsA, history of biologicals and DMARDs. Presence of risk allele for each polymorphism in shown. Upper bars indicate the phenotype of patients at 12 and 6 months; responders and non-responders patients considering absolute PASI≤1 as effectiveness parameter. (C) Receiver operating characteristic curve for prediction of achieving an absolute PASI≤1 at 12 months based on HTR2A, LCE3D and SPATA polymorphisms, weight, PsA, history of biologicals and history of DMARDs.
Although brodalumab was approved by the European Medicines Agency (EMA) for plaque psoriasis treatment in 2017, there is still a lack of thorough studies assessing polymorphisms as treatment response predictors.4 Our study fills this gap by identifying a set of polymorphisms that correlate with brodalumab response. The collective analysis of these polymorphisms could provide a valuable tool for identifying patients likely to achieve absolute PASI≤1 at 12 months in real-world clinical settings.10
Brodalumab use is approved for the treatment of moderate-to-severe plaque psoriasis in the United States, the European Union, Canada, and certain Asian countries. Although phase II and III clinical trials and real-world evidence confirm its efficacy for the management of PsA,11,12 its approval for this disease is currently restricted to Japan. Remarkably, in our study, the presence of psoriatic arthritis was identified as a risk factor for failure to achieve PASI≤1 at 12 months. Current evidence supports a significant association of weight and prior biological therapy with reduced response to biologic therapy. Therefore, studies have consistently shown a correlation between higher body mass index and decreased responsiveness to biologic agents, notably adalimumab and ustekinumab.13
In addition, the risk of lack of response is increased in psoriatic patients who have previously experienced therapeutic failure with >1 prior biologic therapies or multiple DMARDs.14 Based on these reports, our study has described the association of weight and past medical history of previous treatments with failure to respond to brodalumab in psoriatic patients in our routine clinical practice
Through analysis of polymorphisms relevant to psoriasis we constructed a model including polymorphisms in genes HTR2A, LCE3D, and SPATA2, adjusted for weight, prior biologic or DMARDs therapies, and presence of PsA capable of predicting optimal brodalumab response (PASI≤1) at 12 months. This model may represent a tool with good capabilities to differentiate between responder and non-responder phenotypes in real-world clinical practice.
SPATA2 (spermatogenesis-associated gene 2) has been recently identified as a contributor to TNF signaling, regulating NF-κB activity and cell death induction.15 Additionally, SPATA2 is a risk factor for psoriasis in both Western and Indian populations.16
Our data suggest that the AG-AA genotype for rs495337 (SPATA2) is protective, i.e. individuals harboring this genotype may be more likely to achieve PASI≤1 at 12 months.
HTR2A codes for one of the receptors responsible for serotonin signaling. Serotonin plays a crucial role in modulating various immunological processes, including chemotaxis, leukocyte activation, proliferation, and cytokine secretion. In the present study, we found that there is an association between the presence of the HTR2A CT-TT genotype and the failure to achieve optimal response to brodalumab at 12 months. This result is consistent with our previous results showing an association between HTR2A polymorphisms and an inadequate response to anti-TNF drugs and ustekinumab.17,18
The Late Cornified Envelope (LCE) gene cluster, particularly cluster 3, is a susceptibility locus for psoriasis and PsA,19 as well as for other inflammatory diseases such as atopic dermatitis.20 Our data suggest that harboring the GT genotype for rs4085613 (LCE3D) could be useful to identify patients less likely to achieve optimal response to brodalumab at 12 months in clinical practice.
In conclusion, this study identifies genetic variations associated with the response to brodalumab, providing potential insights into the identification of potential optimal responders that may facilitate personalized treatment choice. While pharmacogenetic research in immune-mediated diseases is advancing, personalized therapy using genetic markers remains uncommon in clinical practice. Despite limitations like the small cohort size, this study represents the first pharmacogenetic exploration of brodalumab response in psoriasis in the real-world clinical practice. Further research is warranted to improve our understanding of the determinants of treatment response.
FundingThis study was funded by Instituto de Salud Carlos III (ISCIII) via grant PI21/01583 to HF and EDT and was co-funded by the European Regional Development. This investigator-initiated study was supported by a grant from LEO Pharma. LEO Pharma was not involved in planning, recruiting or conducting the study, or in the process of manuscript drafting.
Conflicts of interestH de la F declared to have received a grant from Instituto de Salud Carlos III, during the study; EDT declared to have received a grant from Almirall during the study, and payments from Janssen-Cilag, Leo-Pharma, Almirall, Novartis, Lilly, UCB, Boehringer-Ingelheim and Bristol-Myers unrelated to this work. AST declared to have received payments from AbbVie, Janssen-Cilag, Leo Pharma, Lilly, Almirall, Amgen, BMS, Novartis and Pfizer and nonfinancial support from Janssen-Cilag, Leo Pharma and Novartis unrelated to this work. MC Ovejero-Benito declared no potential conflicts of interest (research support) with Leo Pharma unrelated to this work. P de la C declared to have received consultancy/speaker fees and/or travel expenses and/or been involved in clinical trials sponsored by Abbvie, Almirall, Amgen, BMS, Boehringer, Celgene, Johnson&Johnson, LEO Pharma, Lilly, MSD, Novartis, Pfizer, Roche, Sanofi, UCB unrelated to the submitted work. MLV declared to have received payments from Abbvie, Almirall, Amgene, Boehringer, Celgene, Janssen, Leo Pharma, Lilly, Kyowa kirin, Novartis and UCB unrelated to the submitted work. GR declared to have received payments and nonfinancial support from Abbvie, Almirall, Amgen, Celgene, Janssen, Leo Pharma, Lilly, MSD, Novartis, Pfizer, Roche, Sanofi, UCB unrelated to the submitted work. FAS has been a consultant or investigator in clinical trials sponsored by the following pharmaceutical companies: Abbott, Alter, Aptatargets, Chemo, Cinfa, FAES, Farmalíder, Ferrer, GlaxoSmithKline, Galenicum, Gilead, Italfarmaco, Janssen-Cilag, Kern, Moderna, MSD, Normon, Novartis, Servier, Silver Pharma, Teva, and Zambon, unrelated to this work.
The rest of the authors declared no conflicts of interest whatsoever.